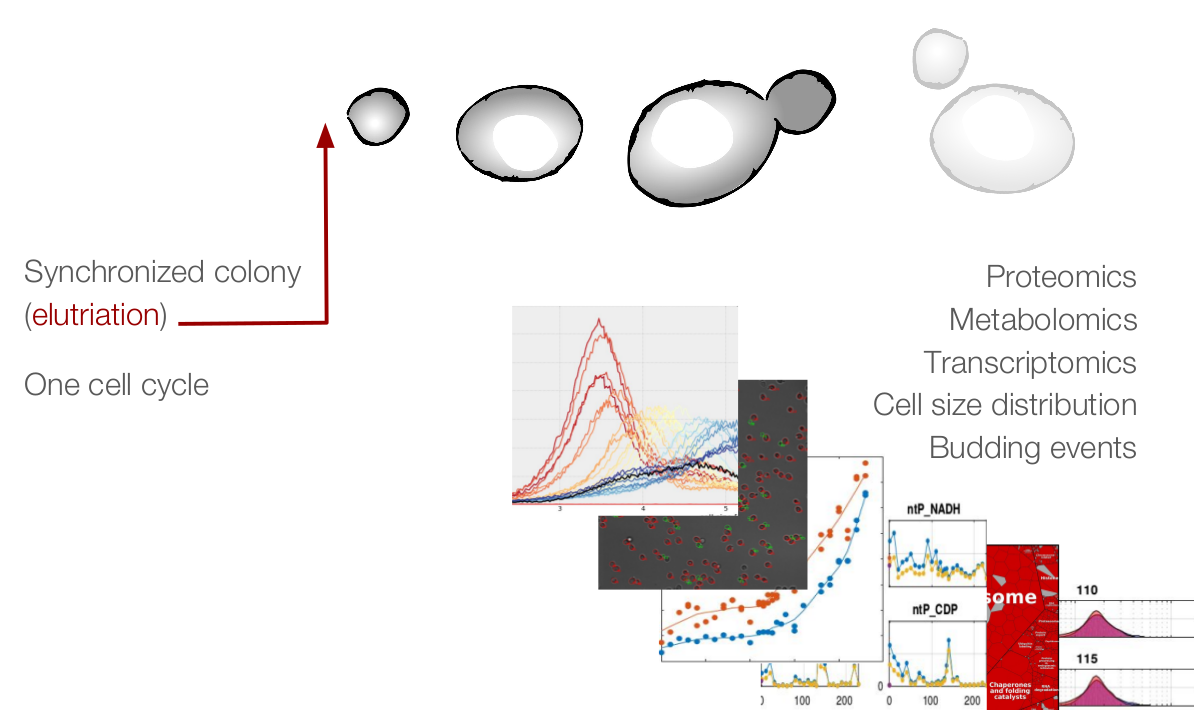
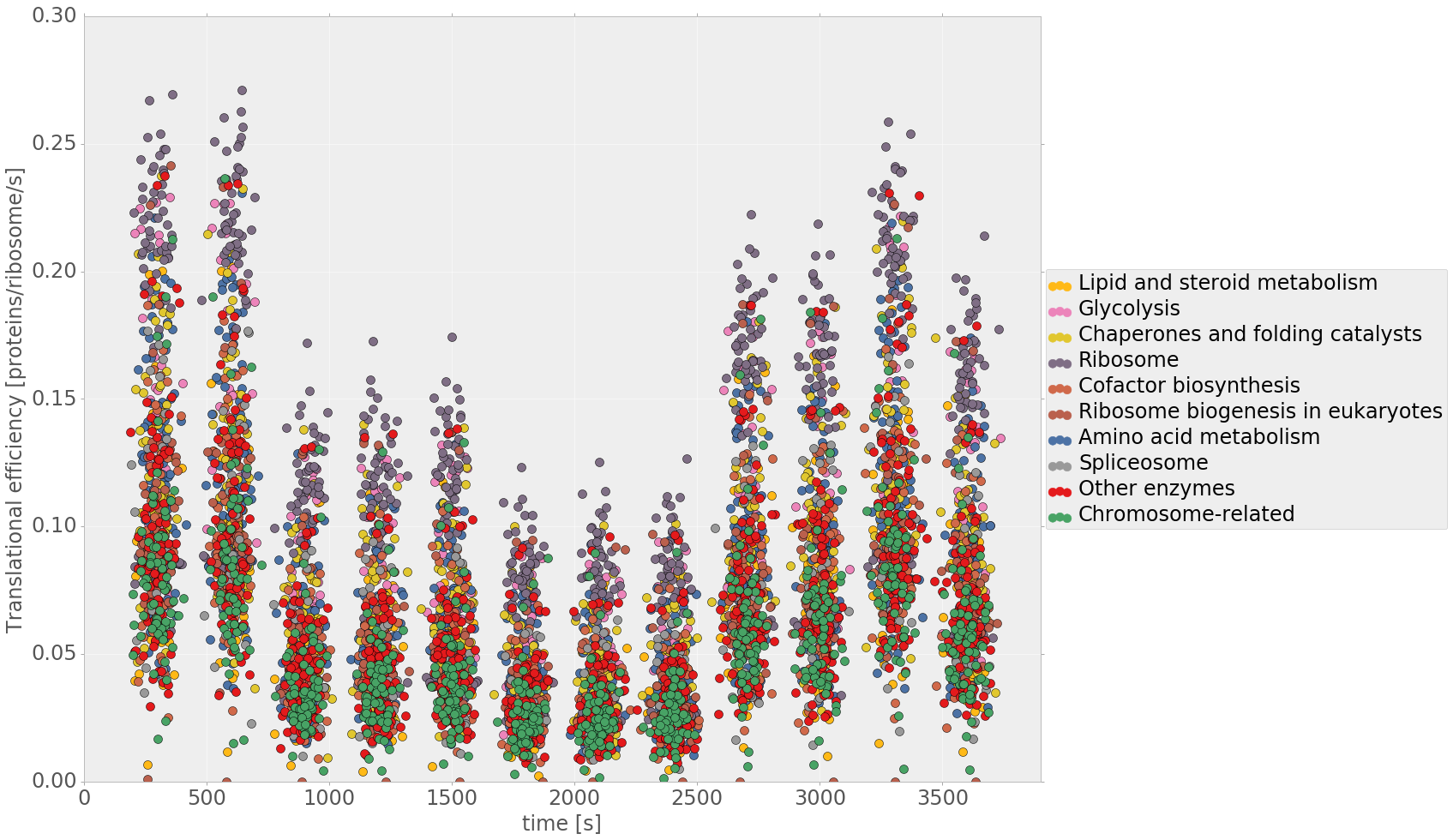
Yeast cell modeling
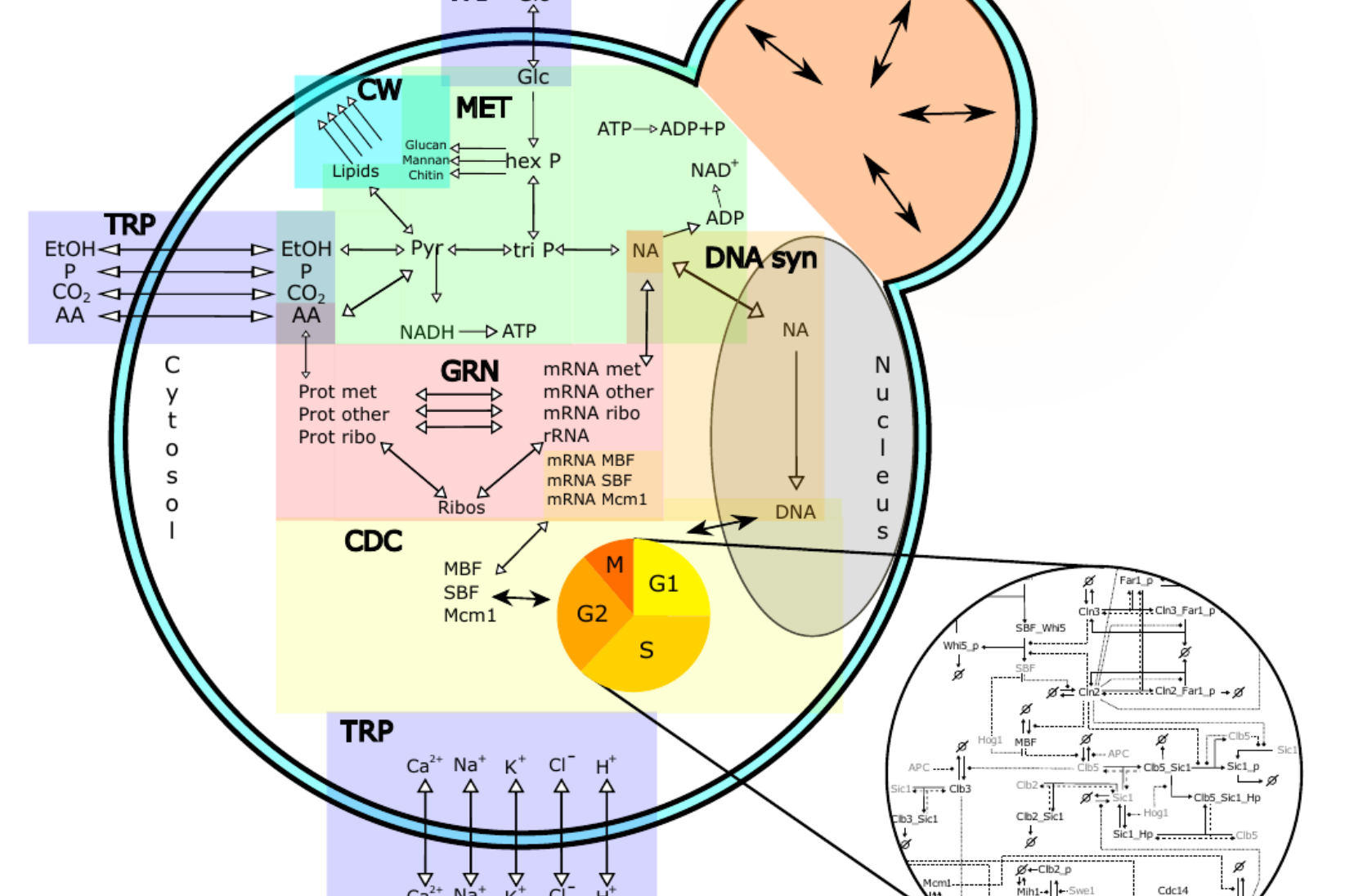
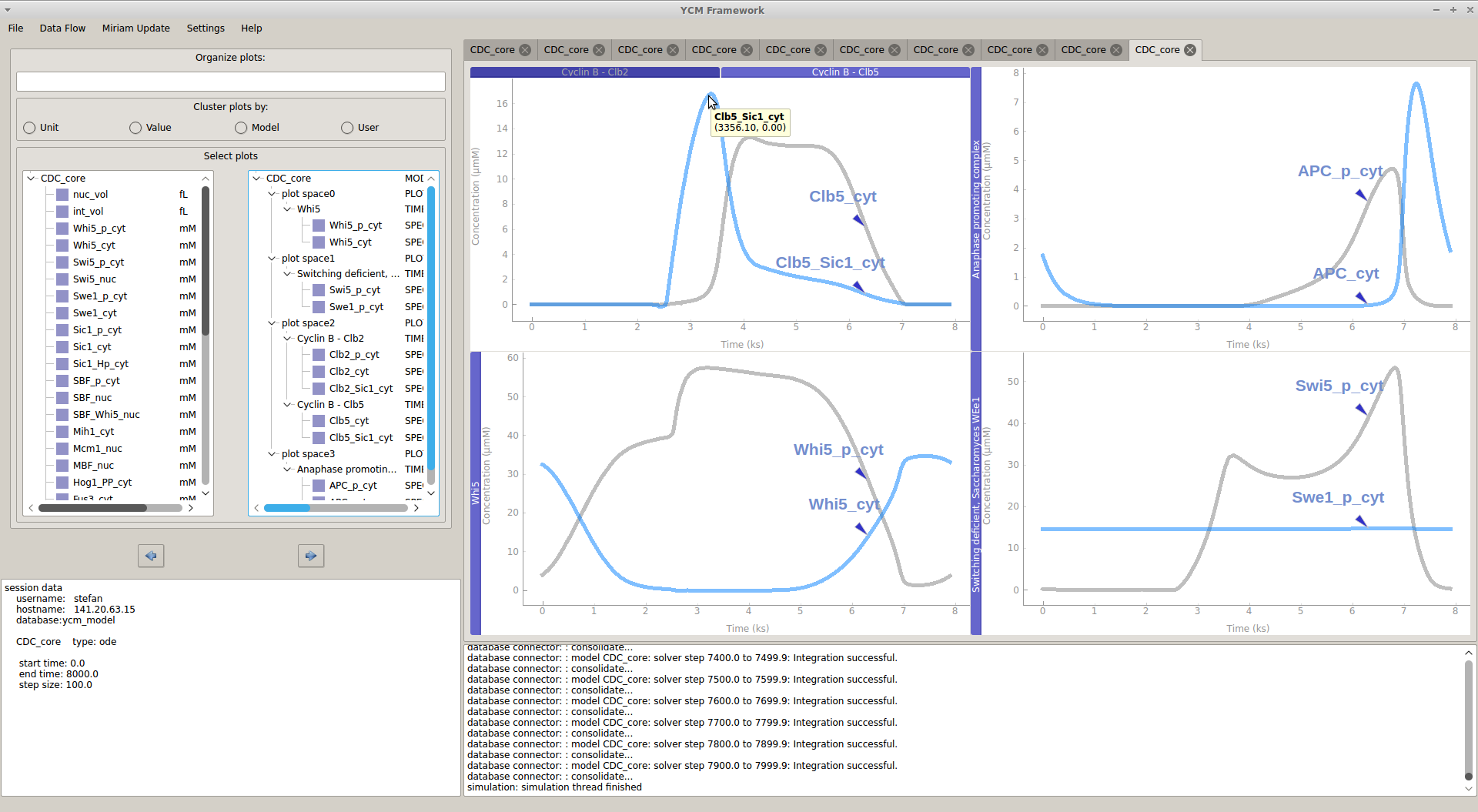
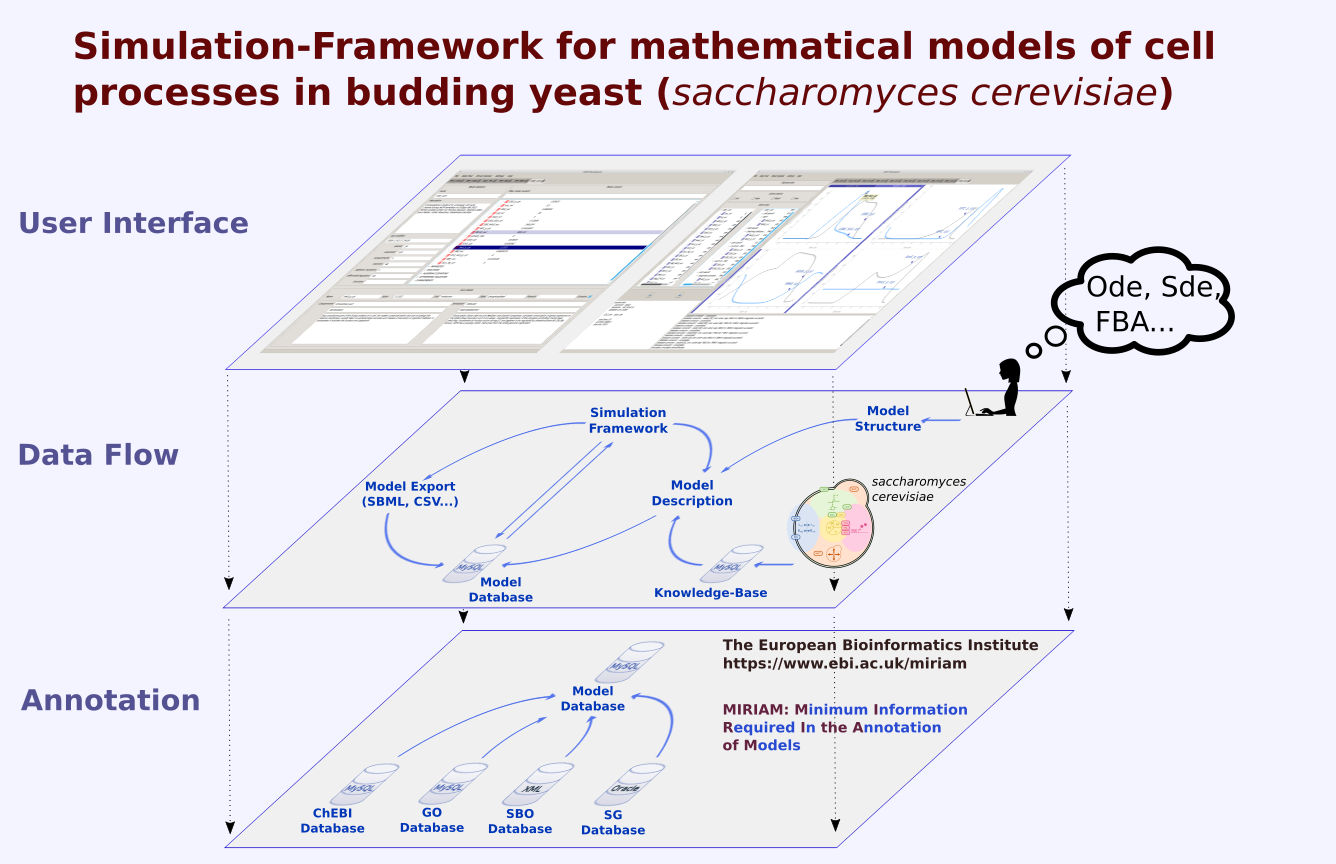
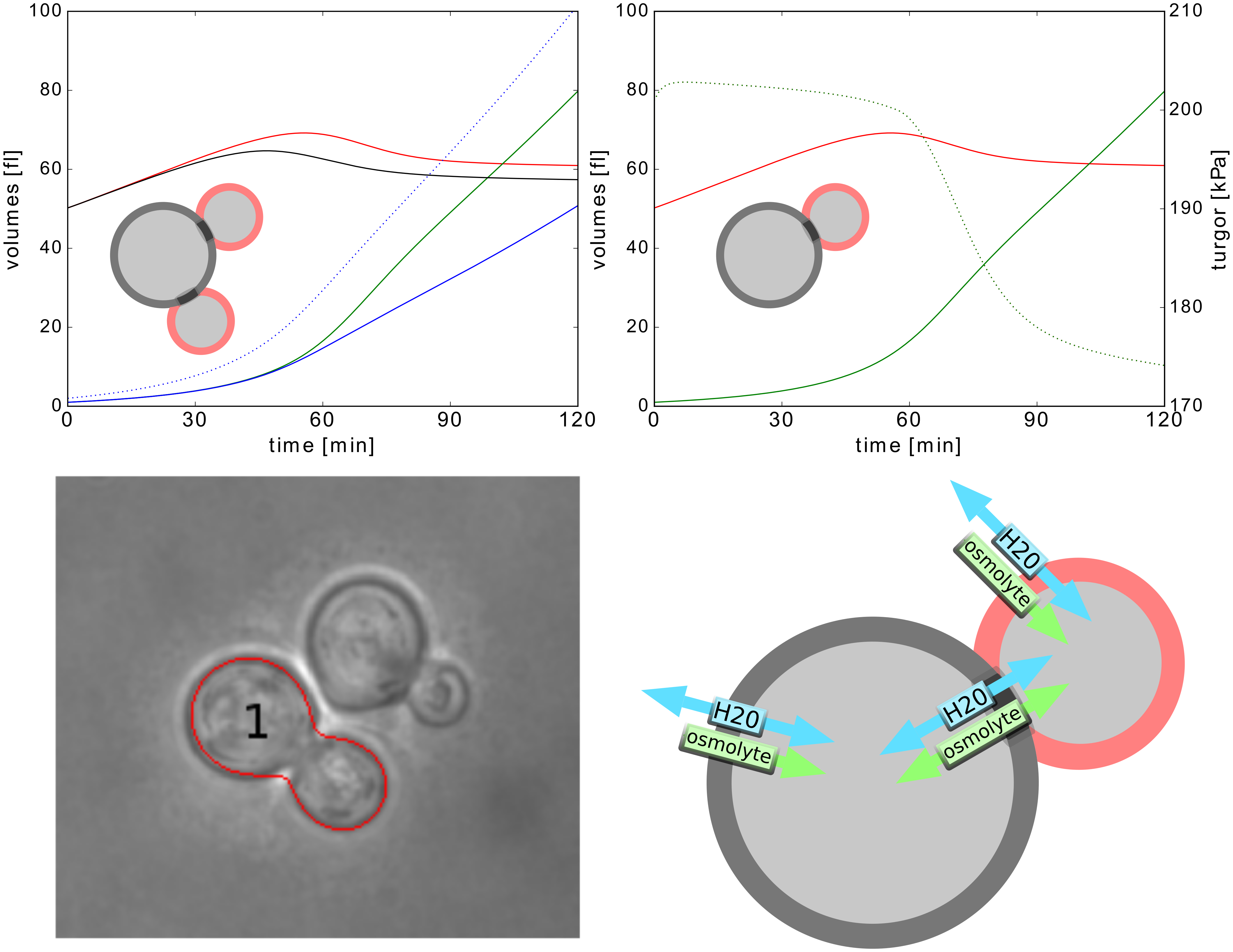
Life of a cell is complex and not only governed by one specific type of network or process. We want to understand how different cellular processes interact to create a functioning cell. How does cell cycle communicate with metabolism? What happens to the translation rate when cellular volume increases? We describe the major processes in yeast during normal growth and upon stress with modular, multi-scale modeling. This allows to understand the systemic connections and mechanisms and explore subsequent effects of cellular perturbations on a systems level. |
|
PEOPLE
PROJECTS
SELECTED PUBLICATIONS
E Klipp, B Nordlander, R Krüger, P Gennemark and S Hohmann.
Integrative model of the response of yeast to osmotic shock.
Nat. Biotechnol. 23 (8):975–982, August 2005.
URLK Tummler, C Kühn and E Klipp.
Dynamic metabolic models in context: biomass backtracking.
Integr. Biol. 7 (8):940–951, 2015.
URLS Gerber, M Fröhlich, H Lichtenberg-Fraté, S Shabala, L Shabala and E Klipp.
A thermodynamic model of monovalent cation homeostasis in the yeast Saccharomyces cerevisiae.
PLoS Comput. Biol. 12 (1):e1004703, 2016.
URLV Schützhold, J Hahn, K Tummler and E Klipp.
Computational modeling of lipid metabolism in yeast.
Front. Mol. Biosci. 3:57, 2016.
URLT Spiesser, C Kühn, M Krantz and E Klipp.
The MYpop toolbox: putting yeast stress responses in cellular context on single cell and population scales.
Biotechnol. J. 11 (9):1158–1168, 2016.
URL
FUNDINGEU - Marie Skłodowska-Curie Innovative Training Network Caroline von Humboldt Professur BMBF DFG
|
COLLABORATIONSDittmar group, Luxembourg Institute of Health (Luxembourg) Sauer group, ETH, Zurich (Switzerland) |