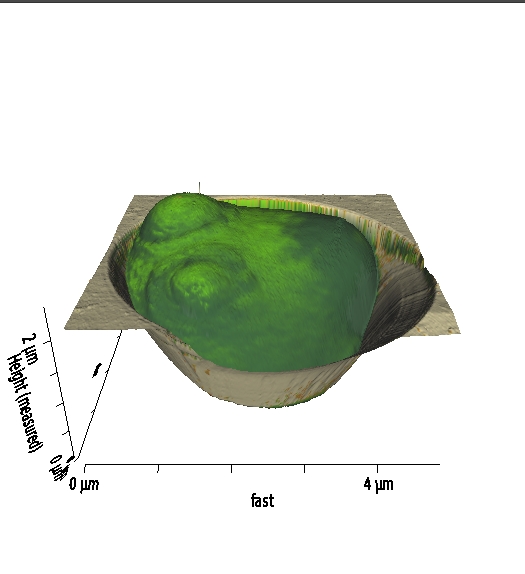
Spatial modeling
How do cells orient themselves spatially in a complex environment? What are the underlying physical mechanisms that cause morphogenetic changes? How do cells break symmetry? How do haploid yeast cells find their mating partner? Using spatial simulation approaches such as partial differential equations or finite element methods, we investigate the dynamics of yeast cells in space and time. |
Image modified from Goldenbogen et al., Open Biol (2016), CC BY 4.0 license |
|
PEOPLE
PROJECTS
SELECTED PUBLICATIONS
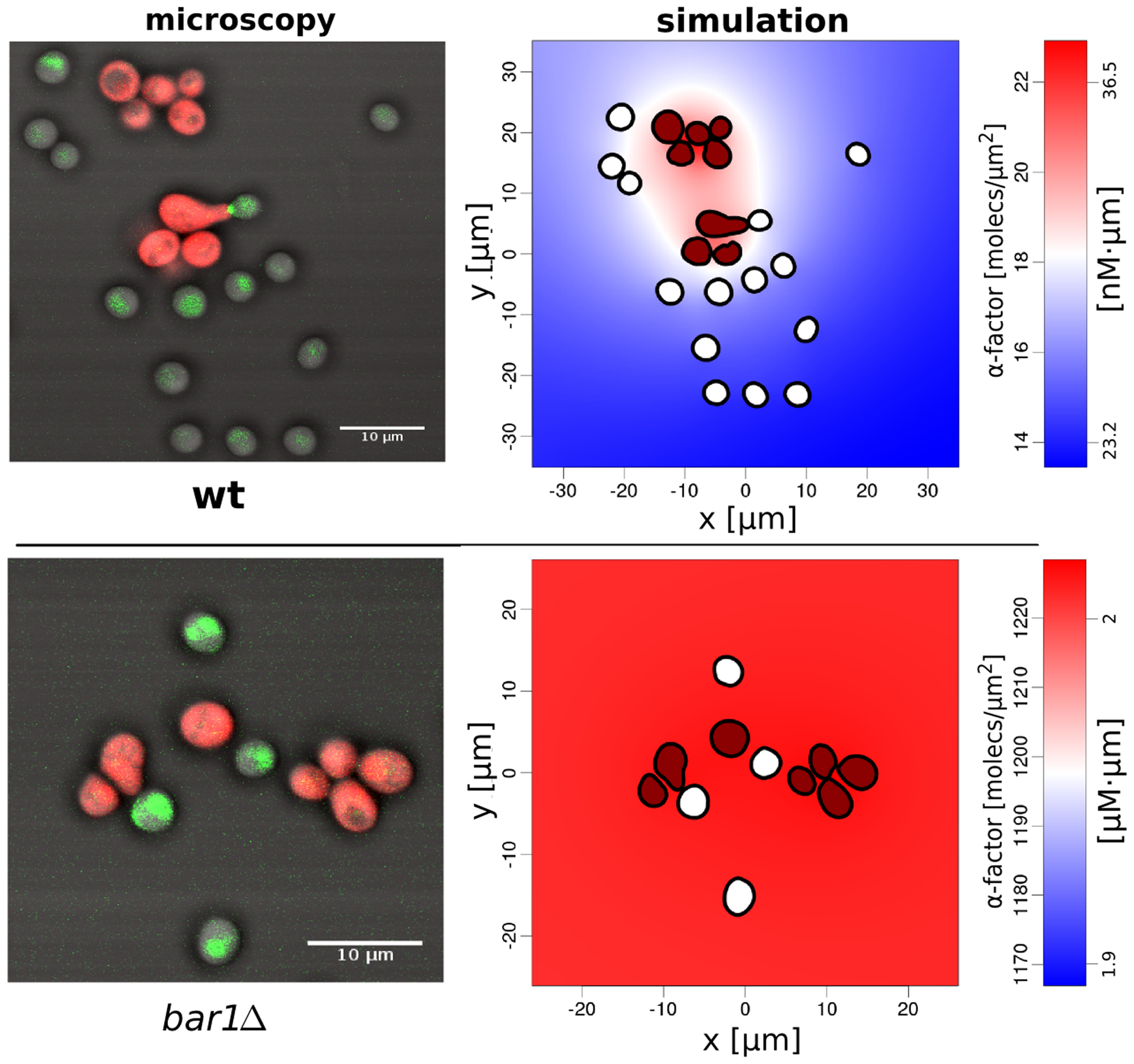
C Diener, G Schreiber, W Giese, G Del Rio, A Schröder and E Klipp.
Yeast mating and image-based quantification of spatial pattern formation.
PLoS Comput. Biol. 10 (6):e1003690, June 2014.
URLW Giese, M Eigel, S Westerheide, C Engwer and E Klipp.
Influence of cell shape, inhomogeneities and diffusion barriers in cell polarization models.
Phys. Biol. 12 (6):066014, 2015.
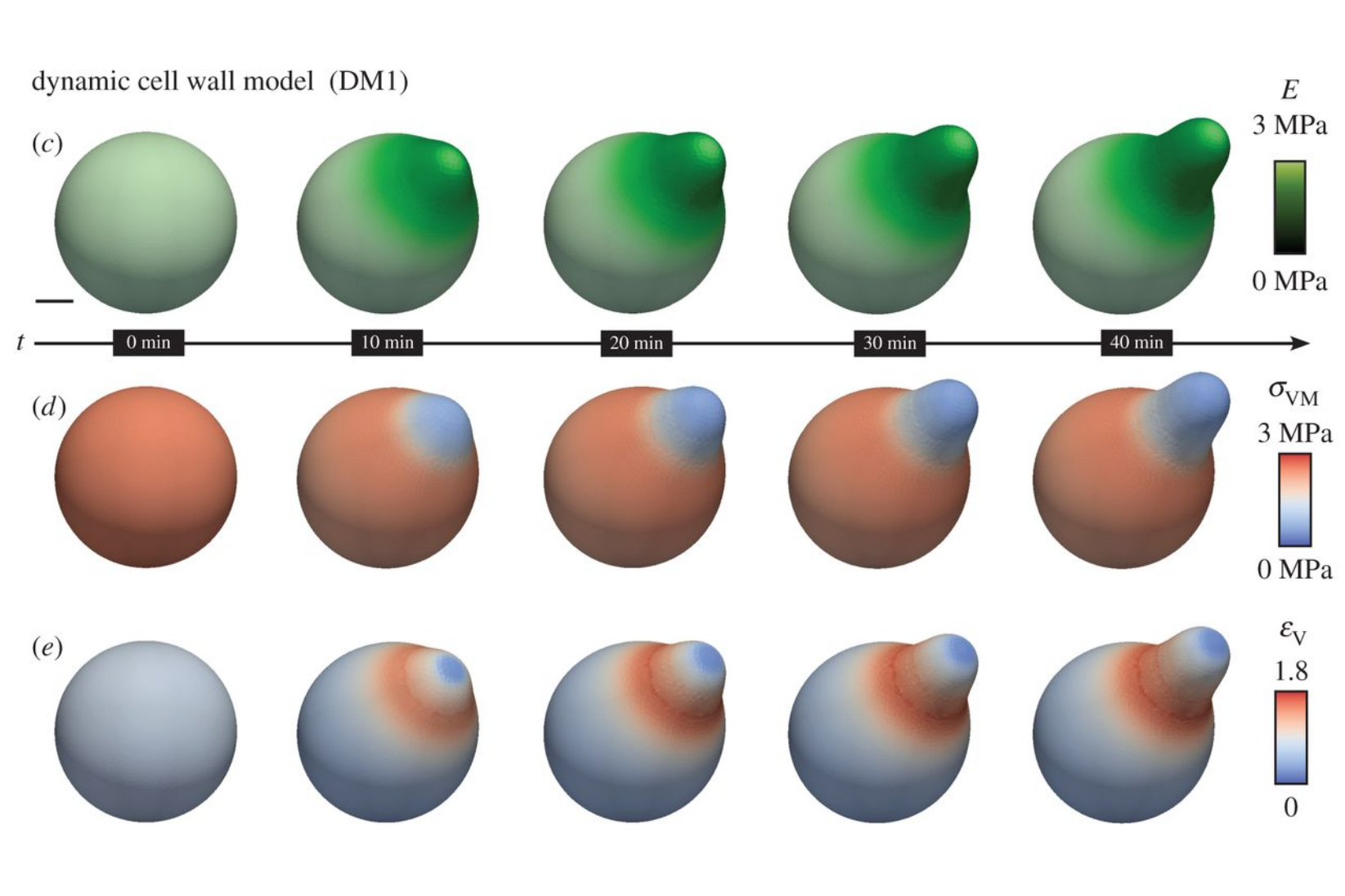
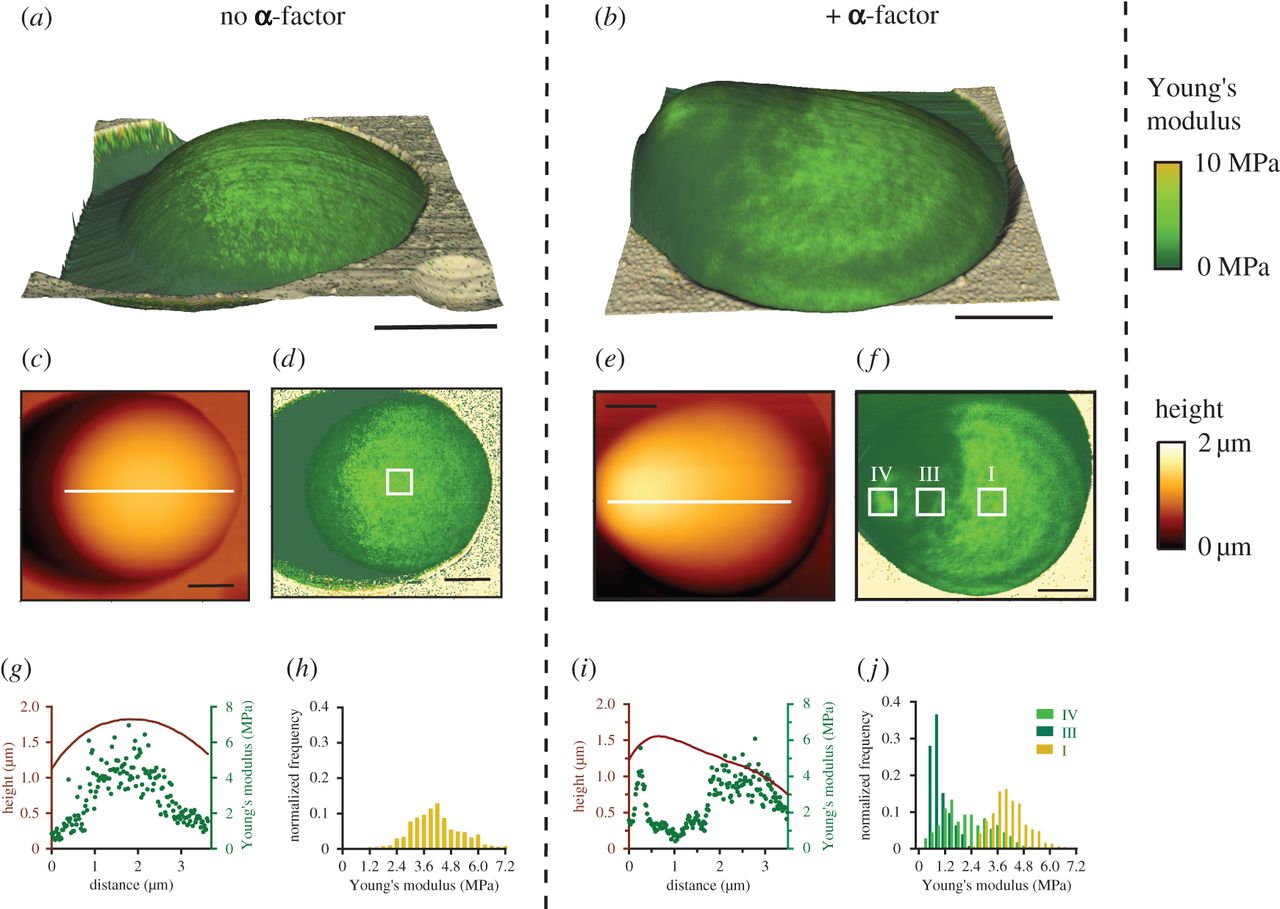
URLB Goldenbogen, W Giese, M Hemmen, J Uhlendorf, A Herrmann and E Klipp.
Dynamics of cell wall elasticity pattern shapes the cell during yeast mating morphogenesis.
Open Biol. 6 (9):160136, 2016.
URLGiese W, Milicic G, Schröder A and Klipp E..
Spatial modeling of the membrane-cytosolic interface in protein kinase signal transduction..
PLoS Comput. Biol. 14(4)(4):1-27, 2018.
URL
FUNDINGDFG |
COLLABORATIONSRobert Arlinghaus group, Leibniz-Institute of Freshwater Ecology and Inland Fisheries, Berlin (Germany) Leibniz-Institut für Zoologie und Wildtierforschung (IZW) Berlin (Germany) |